CoNet - Documentation - Tutorial (Chaffron data)
The goal of this tutorial is to infer relationships between the presence/absence data set assembled by Chaffron.
Tutorial files
Chaffron's presence/absence data spanning multiple environments
metadata accompanying Chaffron's data (to set node colors)
vizmap properties file
settings file
Short version (using the CoNet settings file)
- Open the CoNet plugin in Cytoscape and click the "Settings loading/saving" button.
- In the Settings menu, click "Select file" and browse the file tree to the settings file.
- Click the "Apply settings in selected file" button and close the Settings menu.
- In the Data menu, click on the top-most "Select file" and browse the file tree to select the Chaffron data file.
- In the Data menu, click the "Metadata and features" button to open the metadata submenu.
- In the Metadata submenu, click the left "Select file" button and select the metadata file.
- In the main menu, click the GO button.
Long version (entering everything manually in the CoNet interface)
- Open the CoNet plugin in Cytoscape and open the Data menu
- In the Data menu, click on the top-most "Select file" and browse the file tree to select the Chaffron data file.
- Set the matrix type to "incidence".
- Click the "Metadata and features" button to open the metadata submenu.
- In the Metadata submenu, click the left "Select file" button and select the metadata file.
- In the text area below, enter the metadata columns contained in the metadata file: lineage/taxon/domain/phylum
- Close the Metadata and Data menus and open the "Preprocessing and filtering" menu.
- In "Output filtering", select "copresences only".
- Close the "Preprocessing and filtering" menu and open the Methods menu, then click the "Incidence methods menu" button.
- In the incidence methods menu, enable the "Hypergeometric distribution" checkbox and enter 1 in the text field.
- In the main menu, click the GO button. The computation takes some time (2 minutes on a machine with 8GB RAM).
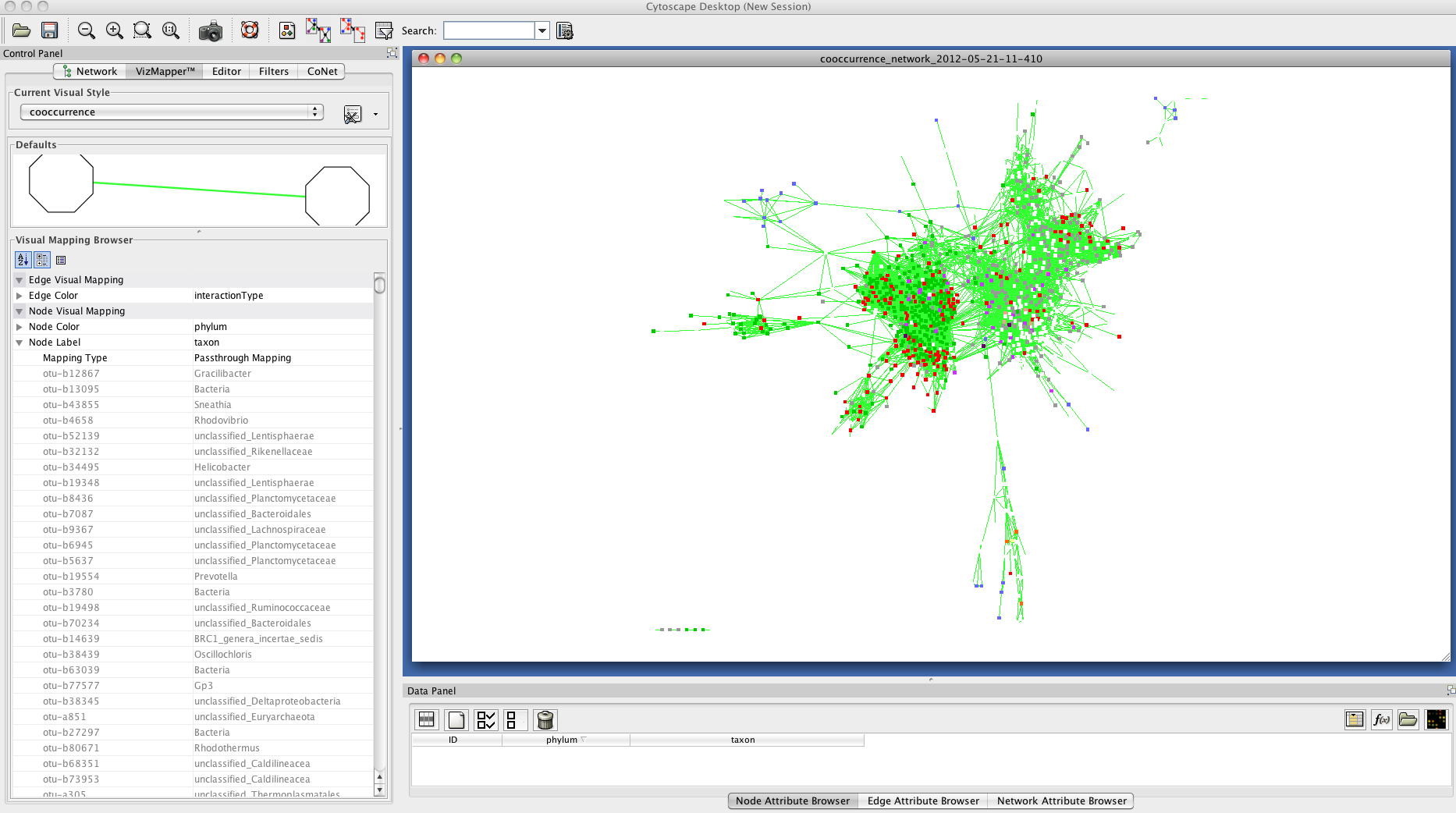
Result
The network returned by CoNet has no layout, as layout computation can be time-consuming, depending on the chosen algorithm.
You can layout the network using one of Cytoscape's supported layouts, for instance the yFiles-layout Organic.
You can then use the VizMapperTM to set phylum-specific node colors or load this tutorial's vizmap property file to set pre-assigned phylum colors. After these processing steps, the network
looks like this:
You can save the current CoNet setting by clicking on the "Settings loading/saving" button in the main menu and then selecting a folder and typing a file name to which to save the current settings.